Background: L-asparaginase (ASNase) converts Asn to Asp and at sustained high levels depletes circulating Asn, leading to leukemic cell death. This dependency has led to the use of ASNase (in a peglyated form, PEGASNase) as an important therapy in the treatment of acute lymphoblastic leukemia (ALL) and has improved survival in patients with ALL. ASNase treatment efficacy relies on significant depletion of circulating Asn for sustained periods of time. Therapeutic monitoring is therefore critical to ensure sufficient levels of ASNase activity to maintain Asn depletion. Serum ASNase activity is monitored as a proxy for Asn levels, having an inverse relationship to Asn. The predictors of serum levels of ASNase activity are not clear however with variation in levels within the same patient between doses. The gut microbiome plays a role in human health and disease, producing metabolites that could impact ASNase therapy. To date, the role of the gut microbiome community in impacting serum ASNase activity levels has not been investigated.
Methods: We investigated 12 paediatric ALL patients for which serum ASNase levels were measured (7 days post treatment) for two consecutive doses of PEGASNase and a stool sample was collected between these two doses (17 samples). Change in serum ASNase activity was determined by examining the difference in consecutive serum ASNase levels. Activity was considered to have decreased when change was negative (serum ASNase levels declined from previous measurement). Gut microbial community composition of the stool samples was determined from a portion of the 16S rRNA gene. In addition whole shotgun metagenome sequencing was used to investigate the relationship between microbial ASNase and ASNS genes and changes in serum ASNase levels during treatment. We utilized a Bayesian model to examine the microbial community structure in serum ASNase decreasing (SD) vs increasing (SI) samples. We used Mann-Whitney U test to examine differences in counts of bacterial ASNase and ASNS genes in SD and SI groups. Finally we investigated counts of bacterial ASNase and ASNS genes along with age, gender, disease risk, dose number, serum ASNase level at previous dose and time between stool sample and dose at predicting change in serum ASNase activity levels using regression models after applying lasso reduction.
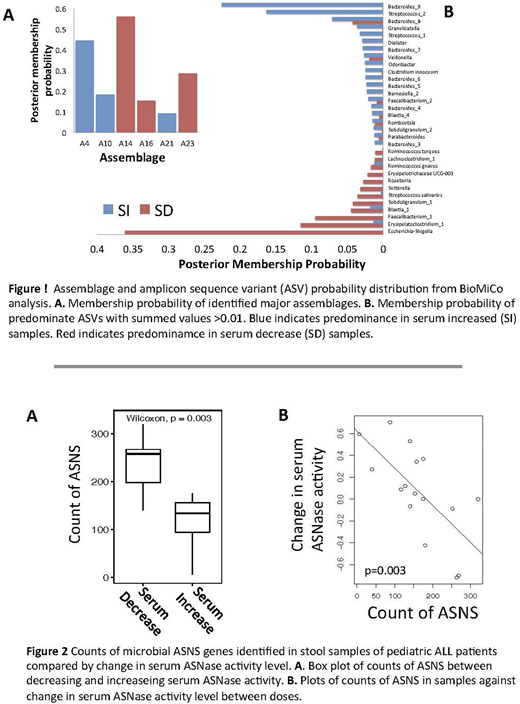
Results: Patients in this study were 50% male and had an average age of 5 years ranging from 1 month to 14.6 years. Among samples examined 35% had decreased serum activity compared to measurements from the previous dose. We identified differing assemblages of microbial taxa prior to PEGASNase treatment. The SD community was predominated by Escherichia prior to treatment while Bacteroides and Streptococcus predominated in the SI community (Fig 1). We found that counts of microbial ASNS were significantly (p=0.003) negatively correlated with change in serum ASNase activity levels (Fig 2), however neither bacterial ASNase gene (ansA or ansB) was significant. Including covariates and applying model reduction we find that ASNS (p=0.0005), dose number (p= 0.001), age at diagnosis (p= 0.001), serum ASNase levels at previous dose (p= 0.008), and counts of ansA (p=0.04) predict change in serum ASNase levels (adjusted R2=0.826, p= 0.0002). Only dose-number was positively correlated with change in serum ASNase level.
Conclusions: We found differences in the microbial community prior to PEGASNase treatment possibly suggesting that modifying the microbiome (decreasing contribution of Escherichia) prior to treatment could result in increased serum ASNase activity. This data also suggests that increased amounts of bacterial ASNS genes present may be associated with a decrease in serum ASNase activity. Future work should focus on a larger and more diverse set of samples in order to further investigate SD and SI community-level properties and the role of covariates (e.g., age and dose number), and further exam the interplay between serum ASNase activity, and bacterial ASNS.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal